This is an article from the fall 2019 issue of LSA Magazine. Read more stories from the magazine.
Anyone interested in using nuclear magnetic resonance (NMR) for their research needs to make sure they leave their car keys, steel-toed boots, cell phones, and credit cards outside the lab. The metal parts could fly across the room and smash into the superconducting magnet inside a big NMR instrument. The NMR’s strong magnet also can disrupt electronics and erase the swipe stripe of credit cards.
But with a few precautions, NMR instruments can safely make some powerful science possible. The magnetic resonance in NMR is the same as in the MRI machines that a patient might find in a hospital. In the same way an MRI can reveal what’s inside a person’s body, an NMR instrument in a science lab can scan a sample to get a sense of what’s inside of it.
An NMR’s enormous superconducting magnets can help reveal the big secrets inside tiny phenomena, such as molecular structure, chemical dynamics, and pharmaceutical targets. “People are using NMR to determine RNA structures for chemistry and biophysics research, or they’re working on large molecules like protein structures,” says Emily Scott, a professor in LSA’s Department of Biophysics, the College of Pharmacy, and the Medical School. “Learning about the structure of a protein or RNA helps researchers find potential drug and disease applications or the basic science of how enzymology works. It helps us understand the dynamics of how these molecules live and breathe, and how they function in the body.”
Scott is the faculty director of the new BioNMR Core on campus, which formally opened in summer 2018 and makes NMR instruments readily available to researchers. For her own research, Scott studies liver enzymes that clear drugs and other foreign chemicals out of the body. “When we determine structures of these enzymes by another technique called X-ray crystallography, we get the equivalent of a snapshot, or a frame in a movie,” she says. “But when we determine the structures of these proteins by NMR, we get to see the equivalent of the movie—how it moves to do what it needs to do.
“It’s much easier to build up a complete knowledge of the functionality of the enzyme if you’re watching the movie than if you’re guessing from a single image.”
LSA runs the BioNMR Core with U-M partners in the Life Sciences Institute (LSI), the College of Pharmacy, and the Medical School. The Core offers equipment that otherwise would be too expensive for most individual labs to buy and maintain. Already, the most powerful NMR instrument in the Core gets booked weeks in advance.
Researchers like David Sherman, a professor in LSA’s Department of Chemistry and LSI, looks at the structure of natural products that he extracts from coral reefs and other natural sites or cultures grown in the lab. His very small quantities require the high sensitivity of instruments available in the BioNMR Core. Chemistry and Biophysics Professor Sarah Keane uses the Core for research aimed at structure determination of large RNA molecules implicated in cancer, heart disease, and bacterial pathogenicity. Anna Mapp, a professor in the Department of Chemistry and LSI; Neil Marsh, a chemistry professor; and members of the Ramamoorthy Lab in chemistry and biophysics all use NMR to figure out the folds in proteins. Their research has relevance to diseases such as Alzheimer’s, Parkinson’s, cancer, and diabetes.
Hydrogen (H) atoms are good to use in NMR, because lots of research samples contain them. Other kinds of atoms can work in NMR, too, including certain kinds of carbon, nitrogen, phosphorus, and more.
Every H atom’s nucleus has one proton. Each proton has a spin and response to magnetic forces.
Each proton acts like a tiny bar magnet, similar to a compass needle, with a north and a south pole.
For these researchers and the students in their labs, the BioNMR Core provides new access to NMR, instrument training, and expertise on campus. Traditionally, these resources had been unavailable to most researchers, with NMR instruments sequestered in individual units or labs. Now, researchers on campus who need to run their experiments with NMR can do it.
The Core comes with an in-house expert, its director Debashish Sahu, who supports faculty research on its NMR instruments and another staff member with expertise in small molecule and protein NMR, Minli Xing. “They’re using newer techniques or even sometimes developing new ways to analyze something that wasn’t possible before,” Sahu says. He helps to create and facilitate those innovations. By staying on top of new NMR technology, Sahu helps researchers take full advantage of the machines for their experiments. “By coming up with intelligent ways of making samples that are visible with NMR, they can tackle problems in ways that no one has done before.” Sahu has the expertise to train new users—including non-specialists — and get those researchers up and running on the instruments.
Because the BioNMR Core centralizes the expensive, highly technical instruments, they’re used efficiently. One lab might run experiments 25 percent of the time, which means the remaining 75 percent can be shared with other researchers. Even off-campus researchers can run experiments on BioNMR Core instruments, which helps to cover operating costs and maximizes the instruments’ use. And Sahu shares his expertise with undergraduate and graduate students in a biophysics course that covers NMR methods.
“Any technique that gives you a glimpse of how biomolecules work that you’ve never seen — that nobody’s ever seen — is really exciting,” says Scott. “That’s why many of us are in this business. For the discoveries.”
NMR In Four Steps
LSA walks you through the process LSA scientists use to research tiny and tough-to-see elements.
1.) At rest, all protons in H atoms naturally orient in random directions in a sample.
2.) In the same way a compass needle aligns with the Earth’s magnetic field, most protons align with a strong external magnet, such as the superconducting magnet in an NMR instrument. Aligning takes the least amount of energy.
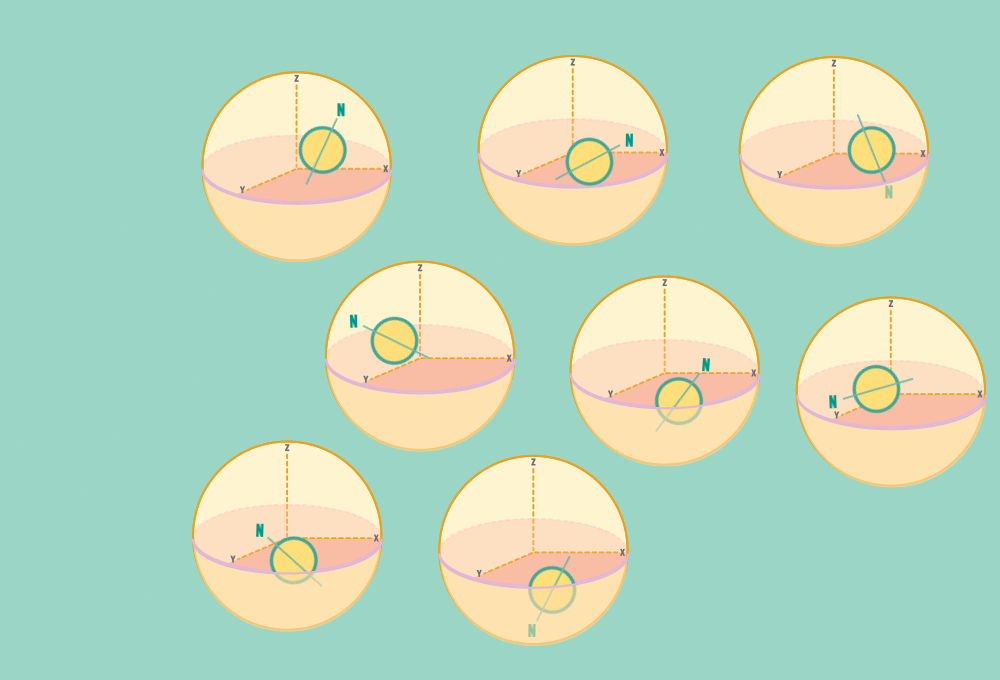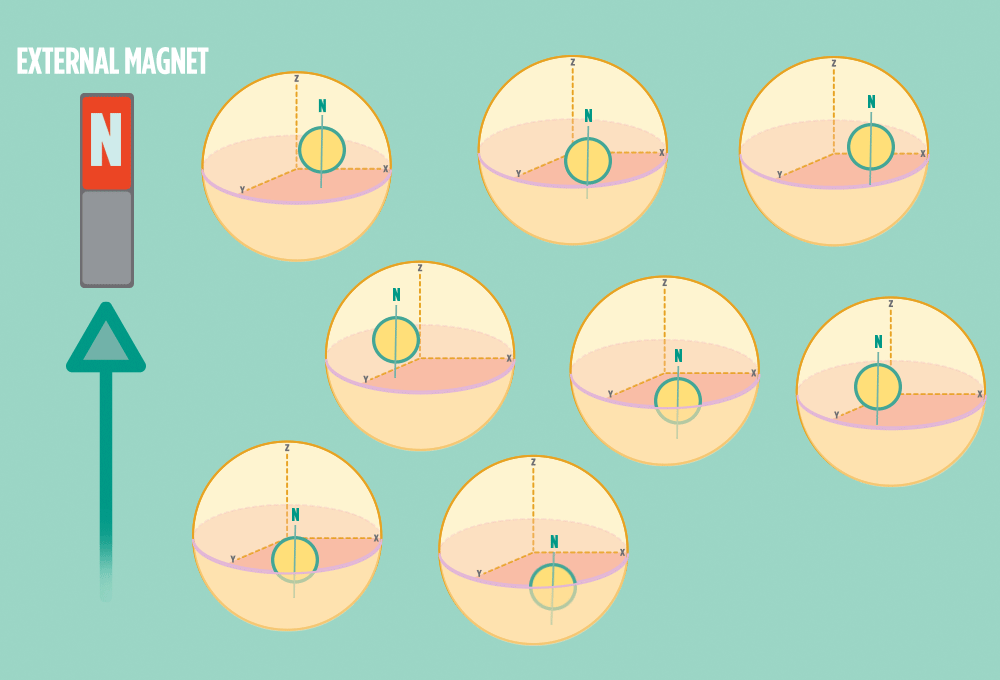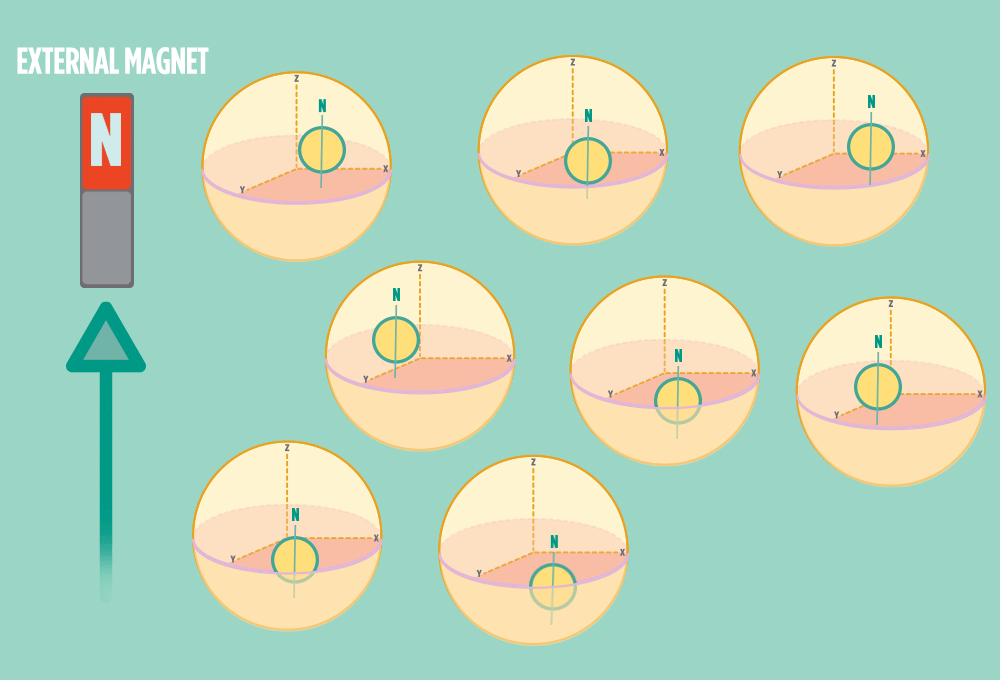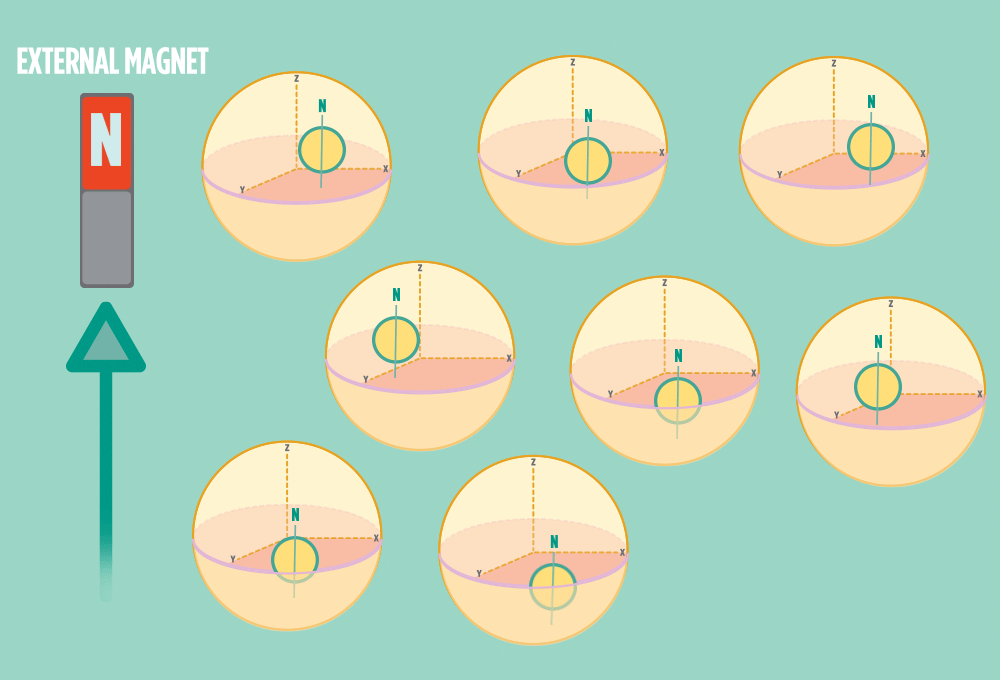
3.) Sending a radio-wave pulse through a sample can add enough energy for the protons to absorb the energy and flip direction.
The amount of energy that gets a proton to flip depends on what surrounds the proton, what’s attached to the proton, and other factors close to the proton.
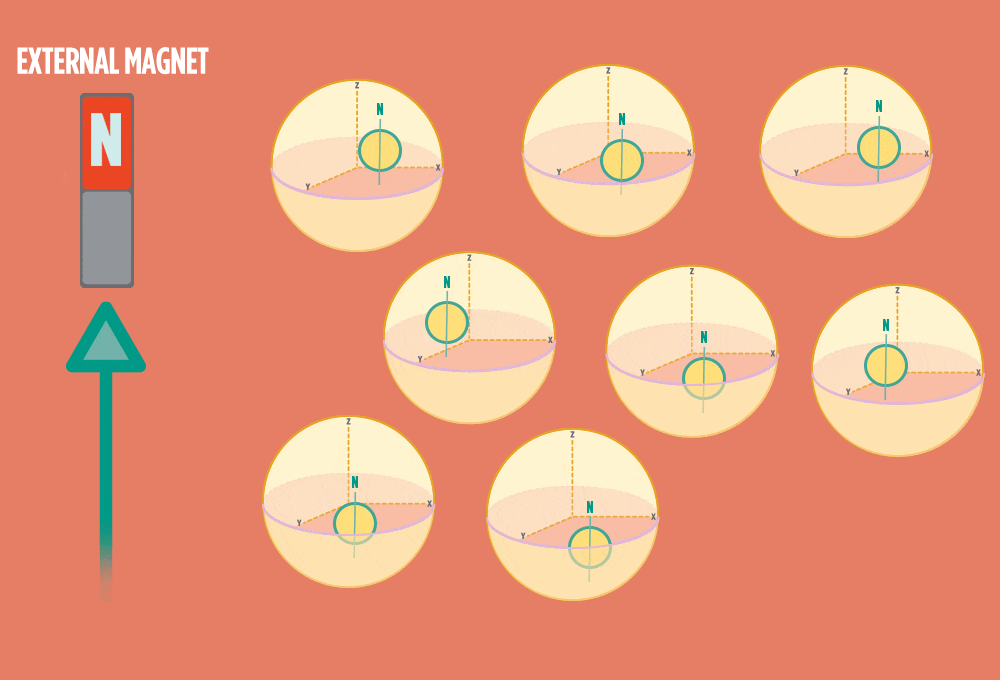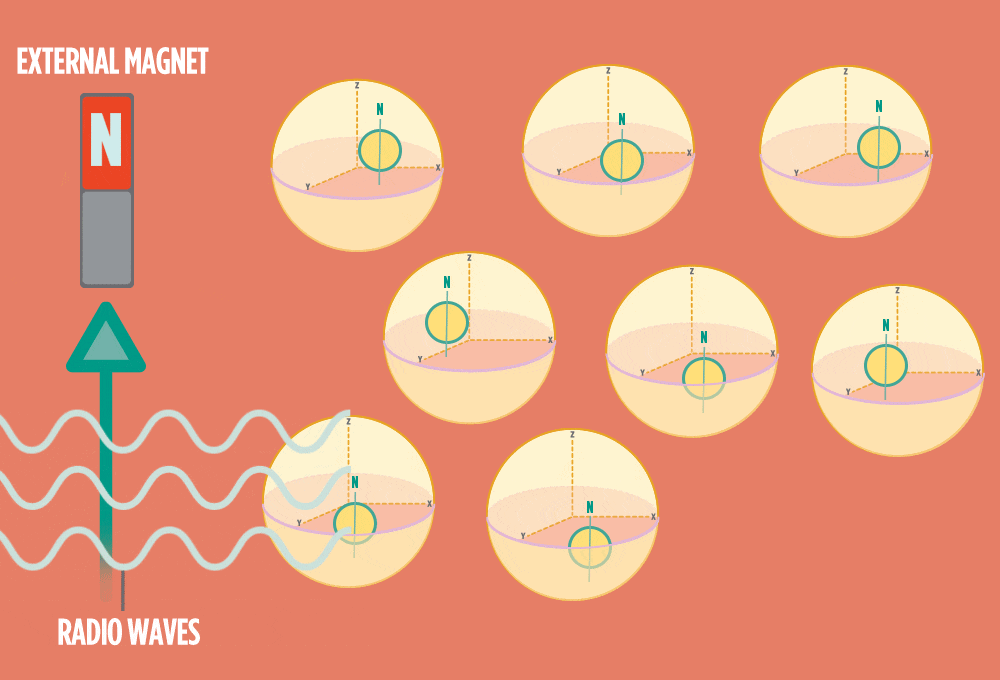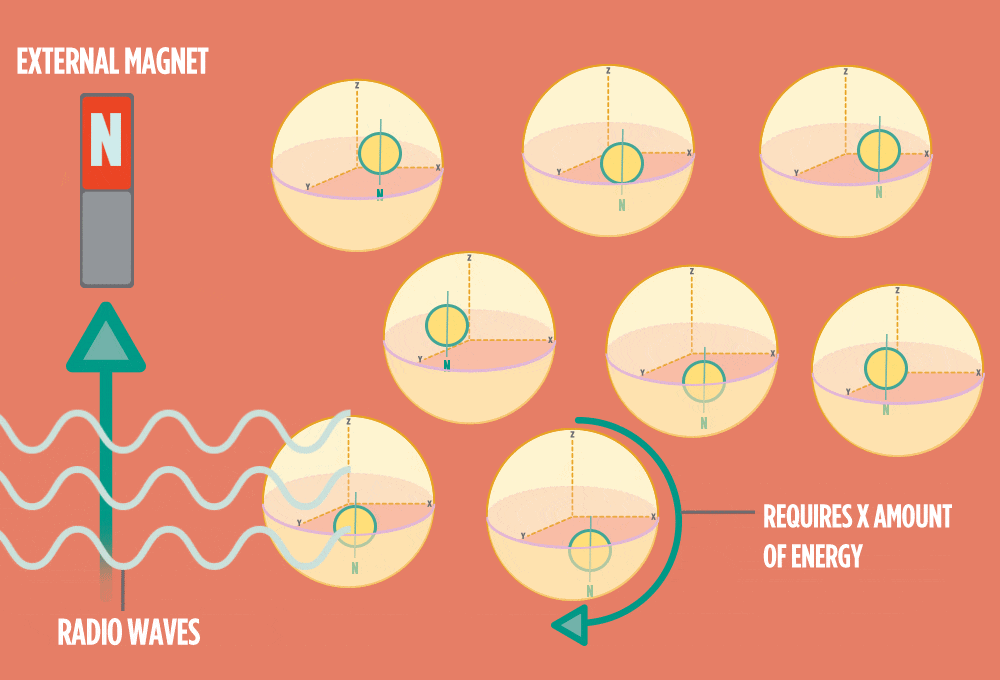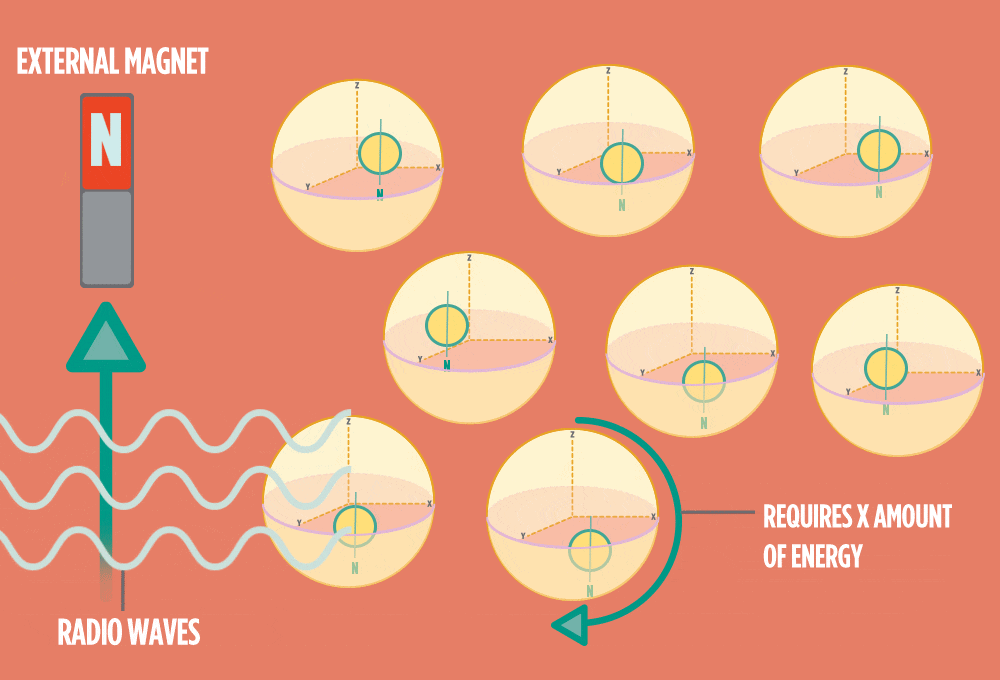
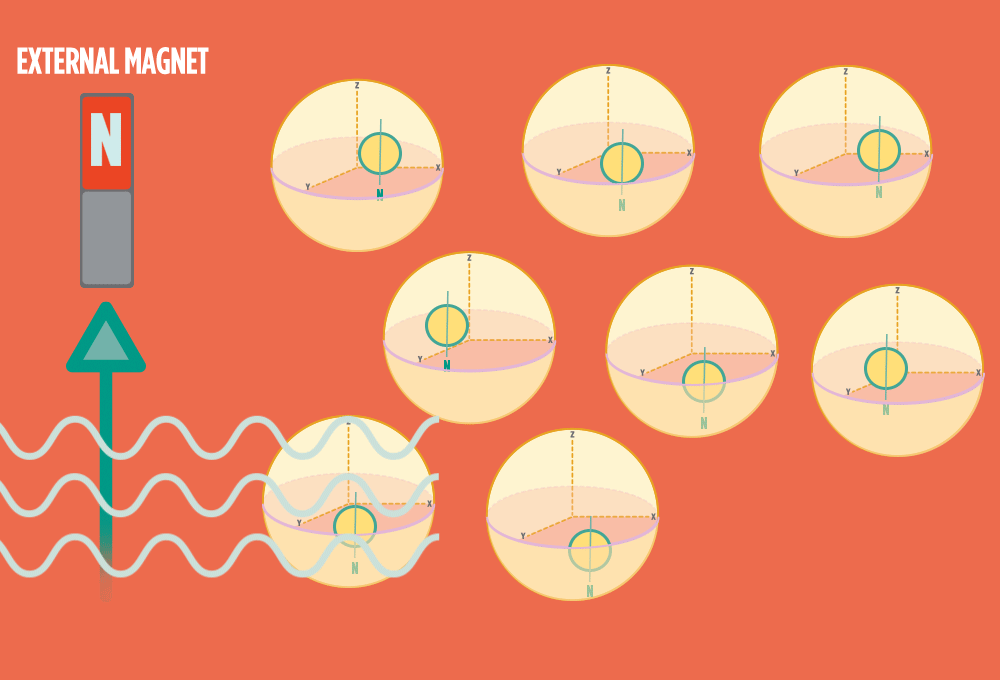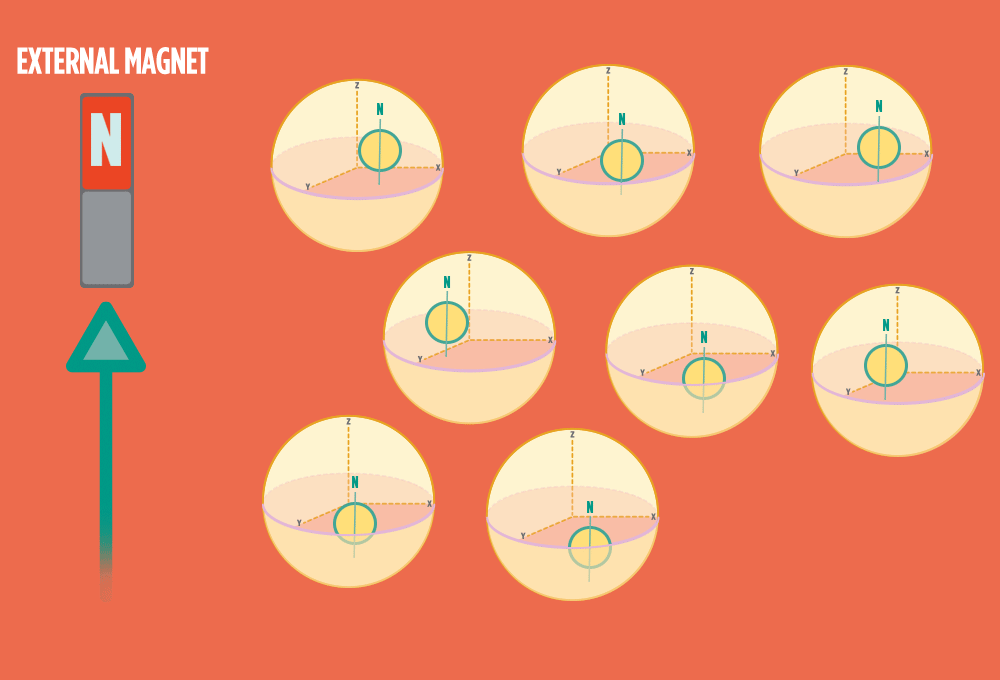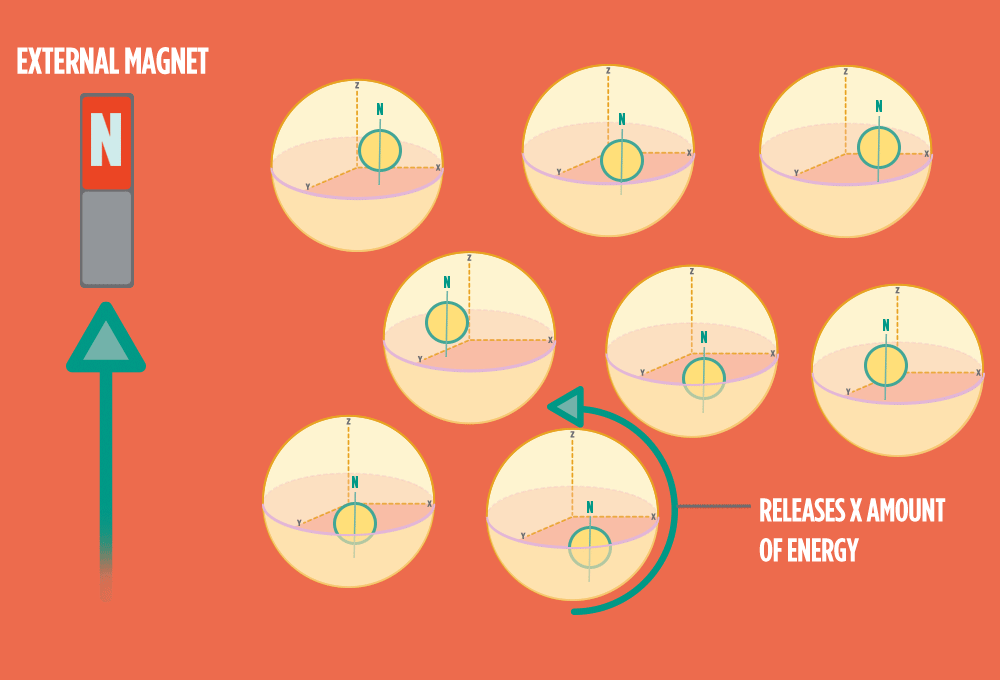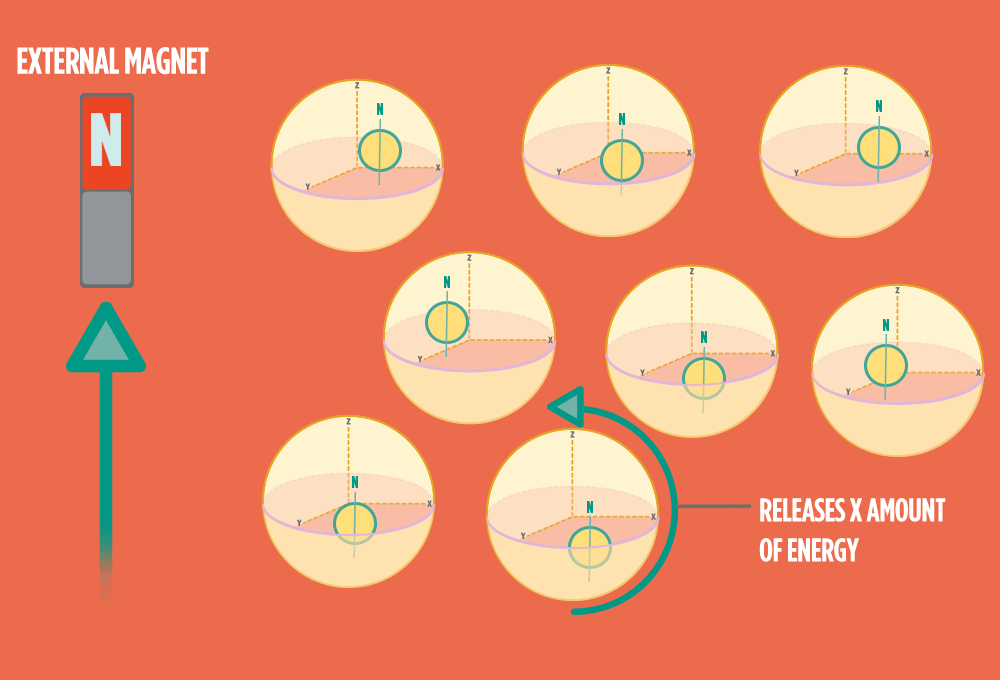
4.) When the radio-wave pulse goes away, protons relax back into alignment with the external magnet, and protons release the same amount of energy that they absorbed when they relax. NMR measures the energy released by the protons. The amount of energy released is a clue about what surrounds the proton, what’s attached to the proton, and other factors close to the proton.